Part:BBa_K1989042
Contents
Triple spytag with mRFP, flash tag and signal peptide SacB
Usage and Biology
Biological material
In the last few years, hydrogens made from natural or synthetic polymers have been investigated due to their extensive application in clinical medicine and synthetic biology. Comparing to traditional biological material, protein-based multifunctional biological material is low-cost, facile and eco-friendly. However, strategies for assembling 3D molecular networks synthesized only by protein molecular remain underdeveloped. The reason why investigating this technology is still tough is lack of protein-based cross linking agents.
Spytag and mRFP
Inspiring from the self-catalysis of isopeptide bond between Lys and Asp in Streptococcus pyogenes fibronectin-binding protein FbaB, researchers split the catalytic domain and obtain two peptide called Spytag(the short one) and Spycatcher(the long one) which are able to form isopeptide bond with the other without any assistant. By fusing Spytag and Spycatcher with functional domains respectively, researchers solve the problem tactfully. In order to using Spytag and Spycatcher system as scaffold, we fused three Spytag spaced by (VPGVG)4 with 6xHistag in N-terminal and another functional protein mRFP in C-terminal.
Signal peptide
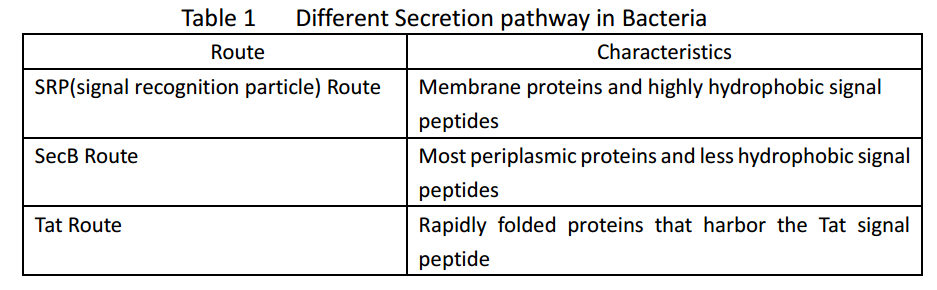
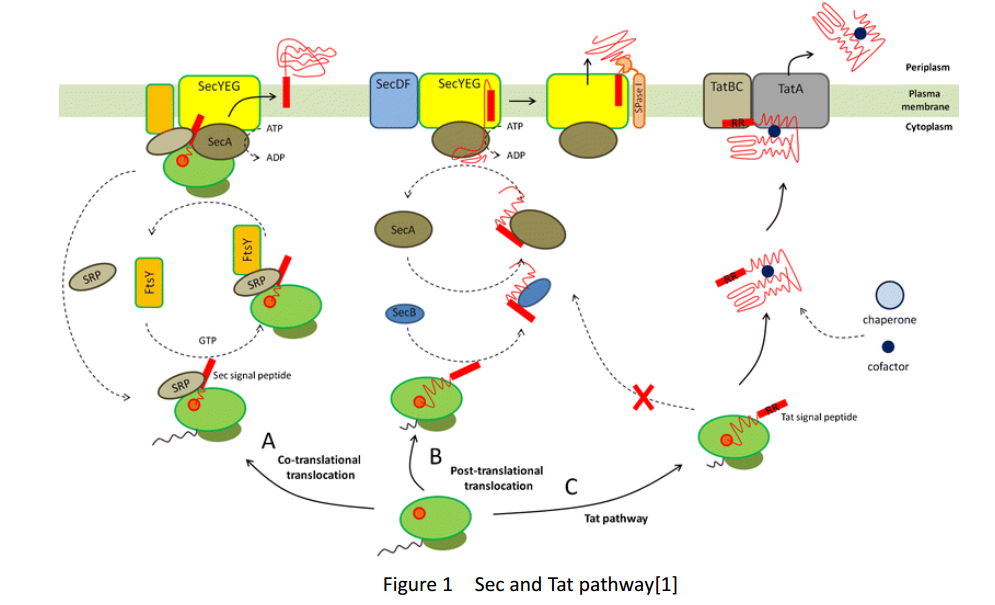
Whether in E. coli or Bacillus Subtilis, the secreted proteins usually contain signal peptides that are essential for export from the cytoplasm. Signal peptides are at the N terminus of target proteins and can be cleaved after secretion. Both E. coli and Bacillus subtilis rely on Sec or Tat machinery to traverse the inner membrane. Detailed description is shown in Figure.1, where three distinct ways are depicted.
As several attributes of our target proteins, such as 3A-SUP, 3B, 3A-mSA, etc. remained unknown, especially their folding state in cells, we were unable to rationally design or choose a signal peptide for each. Nevertheless, we could build a Signal Peptide Repertory to screen for the best SP candidate. Our ultimate goal was to select the most appropriate ones from a huge amount of signal peptides, but owing to the limited time, we had just chosen a limited number of signal peptides to test for its potential to secrete our target proteins. Based on previous studies on their performance of secretion, SacB was selected in this part.
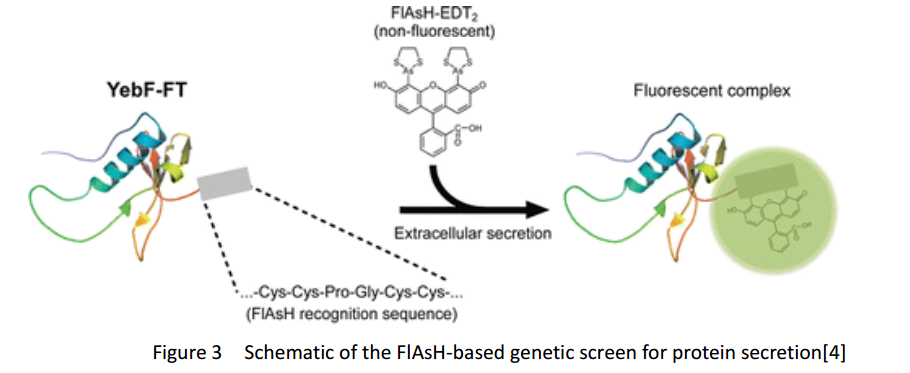
FlAsH-tag
FlAsH-tag is an alias for a tetracysteine-motif-tag (-FLNCCPGCCMEP-) which binds with high affinity and specificity to a biarsenical dye FlAsH-EDT2 and forms a fluorescent compounds.
Based on our results, the fused protein SacB-3A-mRFP-FlAshtag possess both isopeptide bond forming function, coloration ability and secretion . Thus, using Imd-3A-mRFP-FlAshtag as a part of hydrogel formation, we could obtain visible hydrogel.
Cultivation and Result
Cultivation
The part was assembled in pBES plasmid vector Bacillus Subtilis strain harboring the appropriate plasmid was grown at 37 °C in 2xYT medium overnight with suitable concentration of antibiotic.
Result
We could not find detectable protein secretion in the culture medium.
References
1. Low KO, Mahadi NM, Illias RM. Optimisation of signal peptide for recombinant protein secretion in bacterial hosts. Appl Microbiol Biotechnol.(2013)97(9):3811–3826.
2. Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey
3. Hengen, P.N., Purification of His-Tag fusion proteins from Escherichia coli. Trends in Biochemical Sciences, (1995)20(7): p. 285-286.
5. Sun, F. et al. Proc. Natl Acad. Sci. USA 111, 11269-11274 (2014).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 882
Illegal AgeI site found at 1038 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 106
Functional Parameters
| None |